Mai 2016 – in progress
In silico prediction of phage-bacteria infection networks as a tool to implement personalized phage therapy
Motivation
The emergence and rapid dissemination of antibiotic resistance worldwide threatens medical progress and calls for innovative approaches for the management of multi-drug resistant infections. Phage therapy might represent such an alternative. This re-emerging therapy uses viruses that specifically infect and kill bacteria during their life cycle to reduce/eliminate bacterial load and cure infections.
Our approach
The success of phage therapy however relies on the exact matching between both the target pathogenic bacteria and the therapeutic phage. Therefore, having access to a fully-characterized phage library is necessary to start with phage therapy. An essential second step to conceive personalized phage therapy treatments is the capacity to predict the interactions between the target pathogen and its potential phage. To address this, we aim at developing predictive in silico models of phage-bacteria infection networks, using genomic features from sequenced phages and bacteria, and taking advantage of bioinformatics and machine learning techniques.
Using the publicly available information from Genbank and phagesdb.org, we were able to construct a dataset containing +1000 known phage-bacteria interactions with corresponding sequenced genomes. An equal amount of potential negative interactions were added to the dataset by considering the specificity of phage-bacteria interactions. We are currently extracting features from the genomes to build quantitative datasets to train machine learning models. These features include distribution of predicted protein-protein interaction scores, as well as proteins’ amino acids frequency and chemical composition. Future work will focus on the development of ensemble machine learning models to optimize the predictive power of our methodology.
Papers – Posters
Carvalho Leite, D.M., Brochet, X., Resch, G., Que, Y.-A., Pena-Reyes, C.A.; Computational prediction of inter-species relationships through omics data analysis and machine learning. BMC Bioinformatics (to appear). 2018
Poster SIB Days 2018 Student Poster SIB Days 2018
June 20-21, 2018
at the SIB Swiss Institute of Bioinformatics Days held in Biel (Switzerland).
Poster SSM 2017
August 30th to September 1st, 2017
at Swiss Society for Microbiology congress held in Basel (Switzerland).
Poster SIB Days 2016
June 7-8, 2016
at the SIB Swiss Institute of Bioinformatics Days held in Biel (Switzerland).
The specific aims of this project
A. Collection & genome sequencing, assembly and annotation:
- Bacteria.
- Phages.
B. Retrospective in silico prediction of phage-bacteria infection networks:
- Develop a novel machine-learning-based methodology to predict phage-bacteria interactions based on sequence data alone.
- For each predicted interaction, identify the most relevant features underlying it (explanatory power of the methodology).
C. Prospective study on bacterial pathogens to discover new interactions:
- Construct large scale phage-bacteria infection networks.
- In vivo model of Galleria mellonella.
- In vivo validation of new phage-bacteria interactions using animal models of bacterial infections.
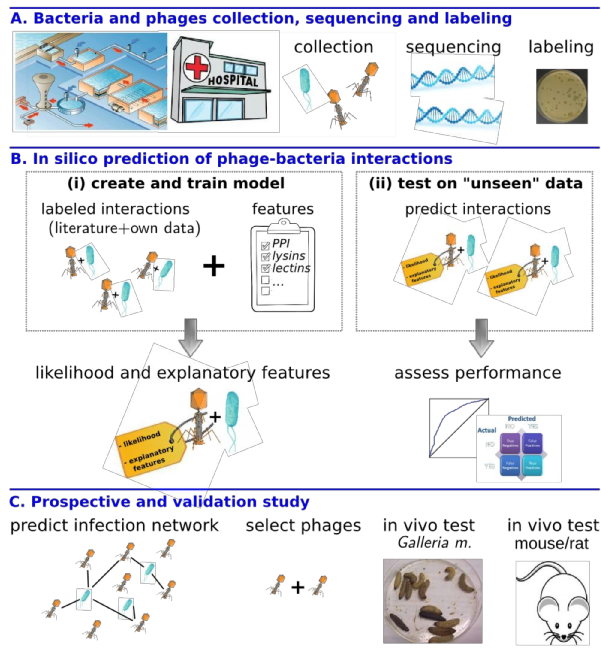 Figure 1: Aims of the project. Phages are represented in orange-brown and bacteria in blue.
Figure 1: Aims of the project. Phages are represented in orange-brown and bacteria in blue.
Collaboration
Dr Grégory Resch Department of Fundamental Microbiology, University of Lausanne, Lausanne, Switzerland.
Dr Yok-Ai Que Department of Intensive Care Medicine, Bern University Hospital (Inselspital), Bern, Switzerland.
