2020 – in progress
Deep Generative Networks for Bacteriophage Genetic Edition
Motivation
Bacterial infections will become highly life threatening events if nothing is done to curb the current trend of antimicrobial resistance. The paucity of potential new anti-infectives in the pipeline of pharmaceutical industries has called for the development of new treatment strategies and incited a renewed interest in bacteriophage (phage) therapy, i.e., the direct use of natural, microbial viruses for the treatment of bacterial infections, as such a potential alternative.
The close evolutionary relationship between phages and their bacterial hosts entails that their genetic information can be used to predict their interactions. The advent of high-throughput experimental techniques paved the way to genome-wide computational analysis and predictive studies. This has motivated several attempts to use bacterial and phage genome analysis, combined with data about known interactions, to train computational models that can predict interactions between new unknown bacteria and/or phages.
Phage therapy efficacy may be hindered by the narrow host range and very high specificity of phages which causes (i) that finding the appropriate phages is a difficult task and (ii) obliging to produce and use cocktails of several phages to fight against simple infections. To solve this difficulty, one forward-thinking modernization of phage therapy is to use genetically-engineered (GE) phages. Such engineered viruses can provide substantial advantages over natural phage in terms of host range, immune system recognition, and environmental stability. A limitation for GE-phages is the empirical knowledge that is currently used to guide genetic modifications. Phages are estimated to be the most abundant organisms on Earth with unparalleled genetic diversity, making it impossible for basic research techniques to factor in all possible variables for creating the most active GE phages.
New technologies are further needed to accelerate the design-build-test cycle for engineering phages and to make it possible to translate proof-of-concept academic work into real-world use more efficiently. The application of Artificial intelligence (AI) to this context has potential both to increase the speed at which genomes can be engineered and to enhance the activity of resulting phages. The only applications of AI to phage biology concern mainly predicting species- and/or strain-level interactions, host species prediction, or the identification of novel viral sequences, making the idea proposed in this project highly innovative.
Our approach
This project, PERPHECT, focuses on the combined use of Deep Neural Networks, that enable extracting patterns directly from DNA sequences which are relevant to predict phage-bacteria interactions, and Deep Generative Adversarial Networks, that have the potential to create sequences, very similar to naturally occurring ones. The sequences generated with this approach are intended to maximize phage interaction with a target bacterium, while minimizing potentially-perturbing, length-associated effects such as noise, repeats, and non-informative code. Such essential segments are, then, integrated into selected, existing, phage genomes thanks to a genome-editing strategy based on sequence similarity and alignment. Finally, the resulting engineered phage genomes are evaluated in silico by means of several interaction-prediction computational models so as to validate their potential interaction.
The specific aims of this project
We plan to address our general research goal through the following specific goals:
- Extract from existing databases (to which we have access) a carefully-selected training dataset, with annotated bacterial and phage genomes combined with data on known interactions.
- Test and improve our existing, unpublished, method (based on DNNs) to predict phage-bacteria interactions with the training data.
- Investigate and customize the architecture of such predictive model so as to be associated with a GAN.
- Using GANs and tailored optimization methods, we will develop a method to automatically generate a relevant phage genome for maximising the phage-bacteria interaction.
- We will then validate, in silico , the adequacy of the methods tested on phage-bacteria pairs known to maximize their interaction with different predictive models.
A schema depicting the workflow proposed for this project is shown in Figure 1. The specific goals are described in more detail in the following subsections.
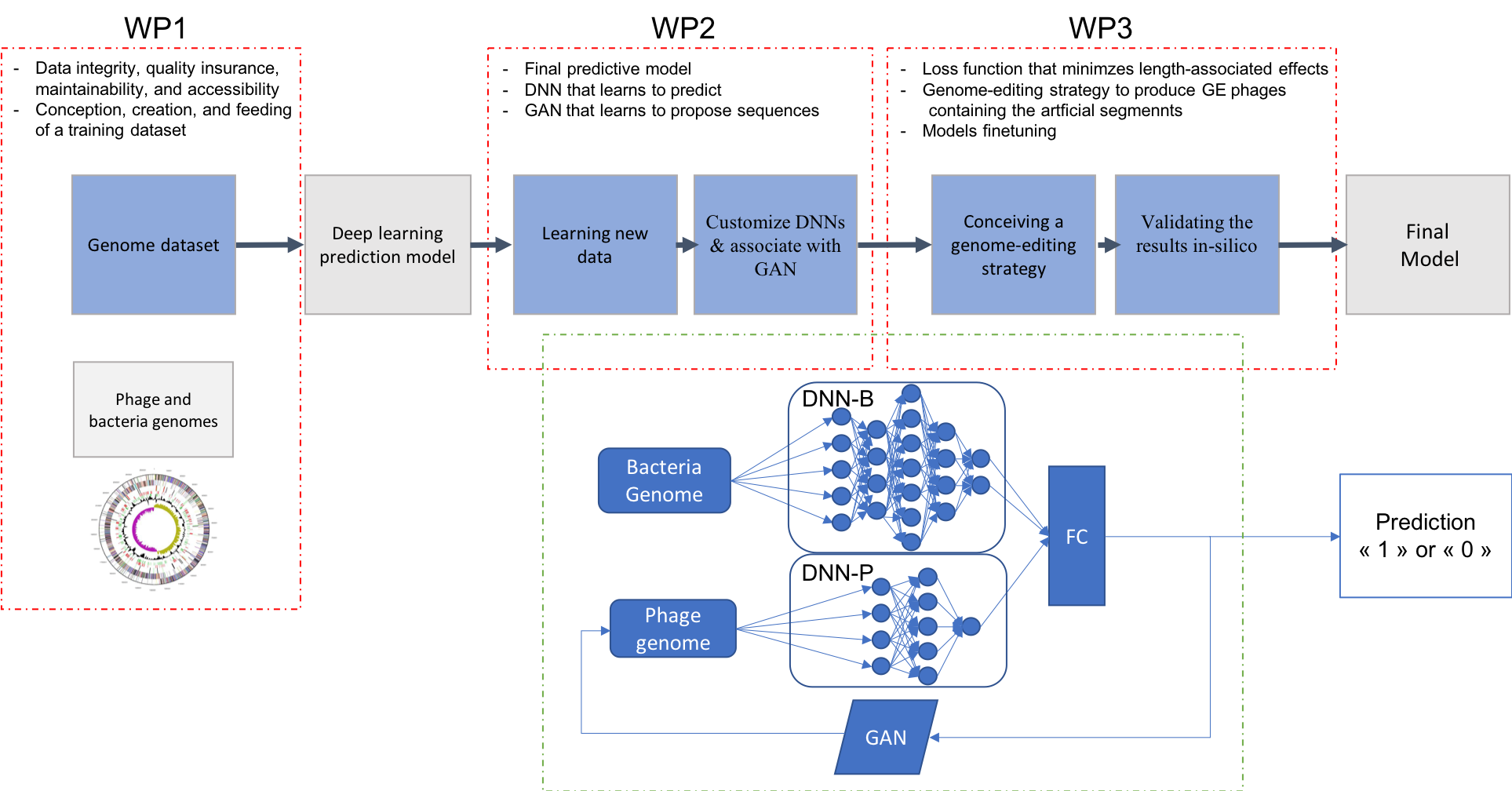
Figure 1: Proposed workflow for the PERPHECT project.
Funding
SNF – Swiss National Science Foundation: Program Spark
