January 2016 – December 2016
“BOSS Explorer” Development and integration of new methods for exploration, selection, and visualization of diagnostic signatures.
Description
SimplicityBio’s competitive advantage lies on its advanced methodology to create and select signatures for diagnostics and drug development, incarnated as a computational tool: the Biomarker Optimization Software System (BOSS). A signature is the combination of a biomarker panel and its classification algorithm. Nevertheless, BOSS is missing a tool for the final selection, navigation and visualization of signatures.
The selection of the final signatures are today performed based on expert knowledge. It takes a lot of time and requires a deep knowledge of fuzzy logic. The usual workflow of this analysis is shown in Figure 1. In addition, the lack of a visual tool increases the time needed to “educate” clients and prospects about the results produced by our platform.
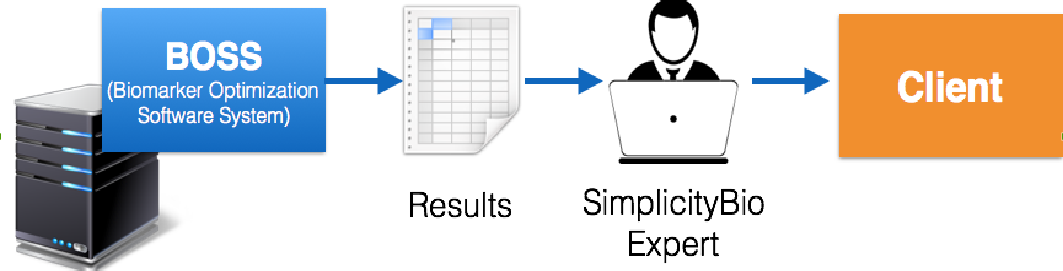 Figure 1. Current workflow for the selection of the final signatures.
Figure 1. Current workflow for the selection of the final signatures.
The proposed project intends to test and implement two main ideas :
1) a visualization tool to improve the exploration, navigation and selection of signatures
2) an extension to the current BOSS methodology to facilitate the selection of the most robust signatures. These two improvements will be marketed as BOSS Explorer.
After several discussions with SimplicityBio’s experts we concluded that the selection of the most relevant signatures can be done in a big part with the aid of a semi-automatic tool. In this context, the expert knowledge can be replaced by new methodologies for selection and visualization of the most relevant signatures. Thus, a tool as BOSS Explorer will remove additional steps, therefore reducing time and expenses (Figure 2).
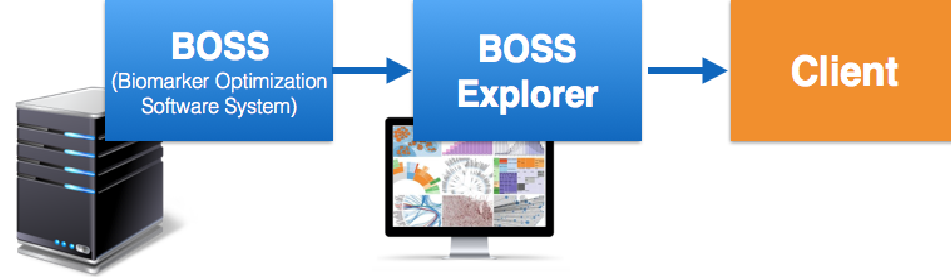 Figure 2. Workflow with BOSS Explorer.
Figure 2. Workflow with BOSS Explorer.
Translating this expert know-how in a semi-automatic tool is a scientific challenge, so we focus this project on two main objectives:
1. To develop a PoC (Proof of Concept) of BOSS Explorer, a tool for selection and visualization of diagnostic signatures.
2. To develop an extension to our current methodology, consisting of new methods to provide optimal strategies to select the most adequate signatures among the candidates and replace expert intervention. It will be focused in the reduction of redundant signatures (signatures using the same panel of biomarkers) and the selection of the most robust signatures.
Partner
SimplicityBio SA (www.simplicitybio.com)
